danRerLib: a python package for zebrafish transcriptomics
1 minute read
While working on my dissertation over the years, I have found some challenges to working with zebrafish transcriptomics data. Most importantly, when looking at biological pathway analysis, there are limited annotations for zebrafish in comparison to other model organisms and especially humans. To make use of the additional annotations out there, we thought of creating a streamlined process for zebrafish researchers to conduct functional enrichment analysis through orthology in Python and release all the code open source through a Python package. Just recently, the associated publication has been accepted in Bioinformatics Advances. If you would like to check out the full publication you can find it here. Below is the link to the comprehensive documentation website for the Python package!

Danio Rerio Libary
danRerLib, short for Danio rerio library, is a comprehensive toolkit designed in Python specifically for zebrafish researchers, focusing on genomics and transcriptomics. Please check out the full documentation for information about the library, installation instructions, and tutorials.
Presenting danRerLib
I have been able to share the work I have done on danRerLib in a couple of different environments so far. More recently, I presented this work at the annual Computational Science Research Center Student Showcase where I was awarded the Windover Ventures Award for my presentation as well as an award for my service to the community.
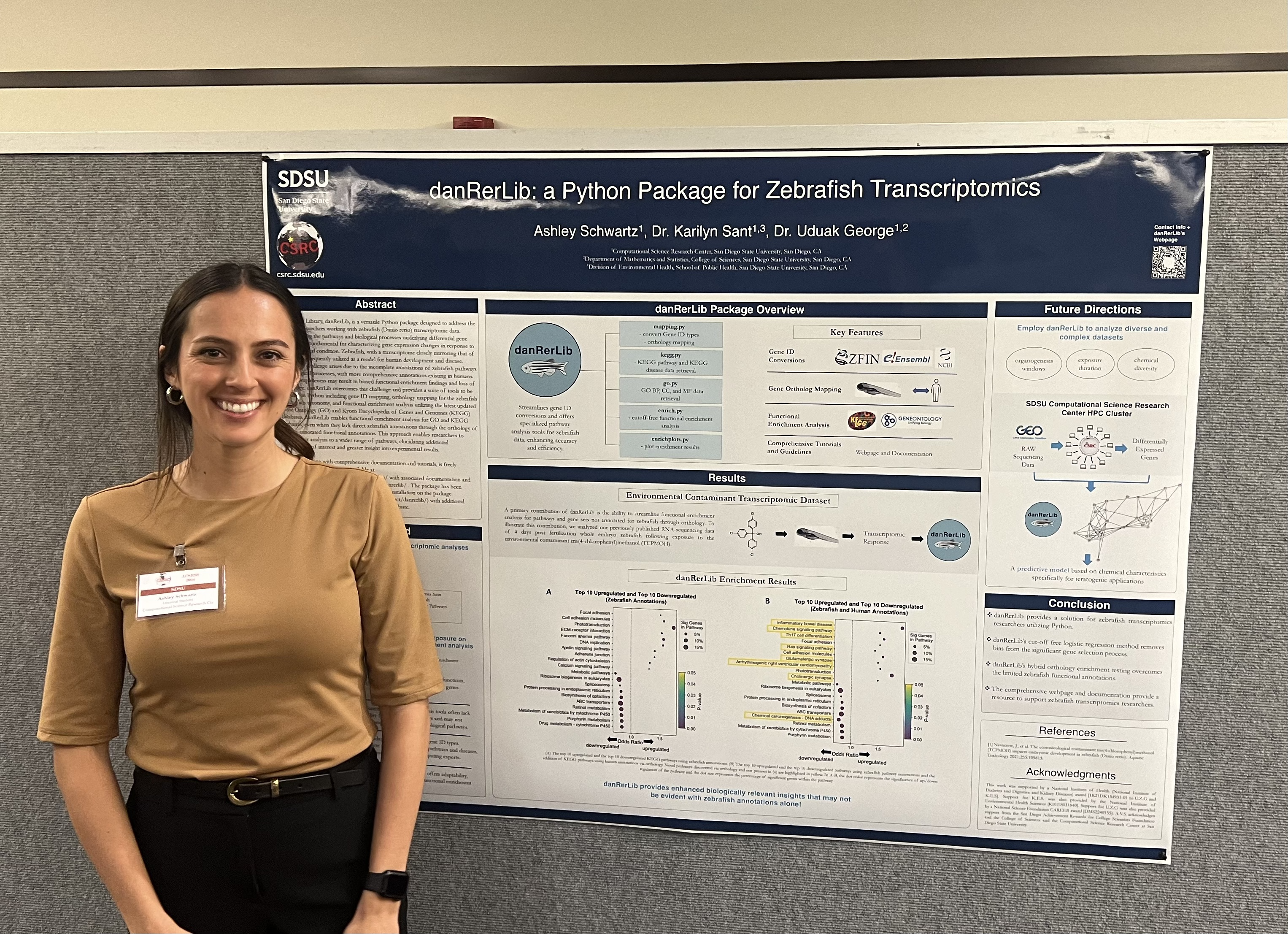
If you would like to learn more about this research and the key findings, you can check out a short quick video I made on the topic.
